Abstract
Introduction: As part of the diagnostic workup in newly diagnosed acute lymphoblastic leukemia (ND-ALL) in adults, chromosomal genomic array testing (CGAT) is frequently added to karyotype and fluorescence in situ hybridization (FISH) to enhance mutational profiling by allowing for more precise analysis of genomic copy number alterations (CNAs) and copy-neutral loss-of-heterozygosity that may impact genes of clinical significance. Despite its increasing use, the prognostic role of CGAT in ND-ALL in adults, and how to apply its findings to clinical decision making, is not well-established. Stanulla and others (JCO 2018) defined the IKZF1plus signature as IKZF1 deletions co-occurring with deletions in CDKN2A/B, PAX5, or PAR1 in the absence of an ERG deletion: this was based on CNAs and carries adverse prognostic significance in pediatric ALL. We hypothesized that by correlating CGAT results at the time of diagnosis with key demographic and outcomes data, we can enhance our ability to risk-stratify patients (pts) at the time of diagnosis.
Methods: Eligible pts included those ≥18 years old diagnosed with ALL at our center between January 2012 and March 2022. We collected relevant demographic, laboratory, treatment, and outcome data. For CGAT, genomic DNA (gDNA) was processed on the CytoScan™ HD Array (ThermoFisher, MA), targeting a genome-wide region with 2.4 million markers for copy number and approximately 750,000 genotype-able small nucleotide polymorphisms (SNPs). The data were analyzed using Nexus Copy Number software with manual curation. Karyotype and FISH were performed in the conventional manner and classified per NCCN. The primary objectives were to characterize recurrent CGAT abnormalities and examine their correlation with survival.
Results: We identified 196 pts with ND-ALL who had CGAT testing performed at diagnosis and received treatment at our center: 54 pts with available data comprised our initial discovery set (Table). CGAT among these pts disclosed large segmental CNAs frequently associated with ALL, such as gains of chromosome X, 1/1q, and 8q; and deletions of 7p, 9p, and 12p. Peak regions with CNAs were detected, which are regions with higher frequencies of aberrations compared with chromosomal segments on either side. We further investigated peak regions that showed aberrations in at least 15% of pts. After removing those that are entirely copy number variation (CNV), the following genomic loci were identified, in order of decreasing frequency: IKZF1 (55%; 17% homozygous loss), CDKN2A (50%), PAX5 (26%), ZCCHC7 (24%), LINC01619 (24%), RB1 (19%), ETV6 (17%), EBF1 (15%), ATP10A (15%), and SLX4IP (15%). The IKZF1plus signature was identified in 30% of pts.
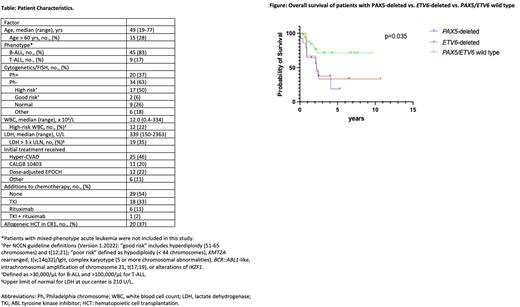
In pts without high-risk cytogenetics (n=17), 88% harbored aberrations in ≥1 of these genes: 29% had IKZF1 deletion and 18% had the IKZF1plus signature. Other commonly observed associations include IKZF1 deletion seen in 85% of Philadelphia-chromosome (Ph)+ pts (n=20) vs. 38% of Ph- (n=34) and CDKN2A deletion seen in 77% of pts with WBC >30K/µL (n=18) vs. 39% without (n=36). With a median follow-up of 2.1 years, there was no significant difference in median OS of the IKFZ1plus cohort (n=17; 4.1 yrs) vs. all IKZF1-deleted (including mono- and biallelic deletions) cohort (n=30; 4.1 yrs), vs. IKZF1 wild type (WT) (n=24, not reached) (p = 0.7). We next created univariate logistic regression models for mortality: statistically significant genomic loci included PAX5 deletion (OR 5.26, 95% CI 1.47-20.0), ZCCHC7 deletion (OR 4.34, 95% CI 1.17-16.67), and ETV6 deletion (OR 5.0, 95% CI 1.07-22.7). In a multivariable logistic regression model for mortality, the strongest associations were seen with PAX5 deletion (OR 4.0, 95% CI 1.0-16.39) and ETV6 deletion (OR 5.0, 95% CI 1.1-27.78): the median OS in the PAX5 deletion (n=14; 2.2 yrs) and the ETV6 deletion (n=9; 2.2 yrs) cohorts was inferior to the PAX5/ETV6 WT cohort (n=35; not reached) (p=0.035; Figure).
Discussion: In this initial discovery-phase analysis of CGAT in ND-ALL in adults, we identified CNAs involving well-described genes (e.g. IKZF1), and lesser-described genes (e.g. LINC016919). Abnormal CGAT was seen in nearly all pts with otherwise normal or good cytogenetic risk. IKZF1plus was not associated with worse survival in this adult cohort, though deletions of PAX5 and ETV6 were. Analysis of additional pts to validate the significance of our findings will be presented.
Disclosures
Percival:Pfizer: Research Funding; Trillium: Research Funding; Oscotec: Research Funding; Cardiff Oncology: Research Funding; Glycomimetics: Research Funding; Biosight: Research Funding; Celgene/BMS: Research Funding; Abbvie: Research Funding; Ascentage: Research Funding; Telios: Research Funding. Ghiuzeli:Seattle Genetics: Current equity holder in private company; Crisp: Current equity holder in private company; Intellia: Current equity holder in private company; Repare therapeutics: Current equity holder in private company; Uniqure: Current equity holder in private company; Axson therapeutics: Current equity holder in private company; TG Therapeutics: Current equity holder in private company; Sangamo therapeutics: Current equity holder in private company; Sorrento therapeutics: Current equity holder in private company; Sorrento therapeutics: Current equity holder in private company. Cassaday:Jazz: Consultancy; Merck: Research Funding; Servier: Research Funding; Vanda: Research Funding; Pepromene Bio: Membership on an entity's Board of Directors or advisory committees; Autolus: Membership on an entity's Board of Directors or advisory committees; Kite, a Gilead Company: Consultancy, Research Funding; Pfizer: Consultancy, Research Funding; Amgen: Consultancy, Research Funding; Seagen: Current Employment, Current equity holder in private company, Other: Spouse employment with Seagen; stock or other ownership in Seagen..
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal